In a groundbreaking study by researchers at the Institute for Molecules and Materials at Radboud University in the Netherlands, the potential of molecular computing has been brought to light. This study, published in Nature and led by Prof. Wilhelm Huck, delves into the intricate world of complex self-organizing chemical reaction networks and their ability to perform a multitude of computational tasks. The field of molecular computing has long been of interest to researchers seeking to unlock the computational power of chemical and biological systems, where chemical reactions act as reservoir computers, transforming inputs into high-dimensional outputs.
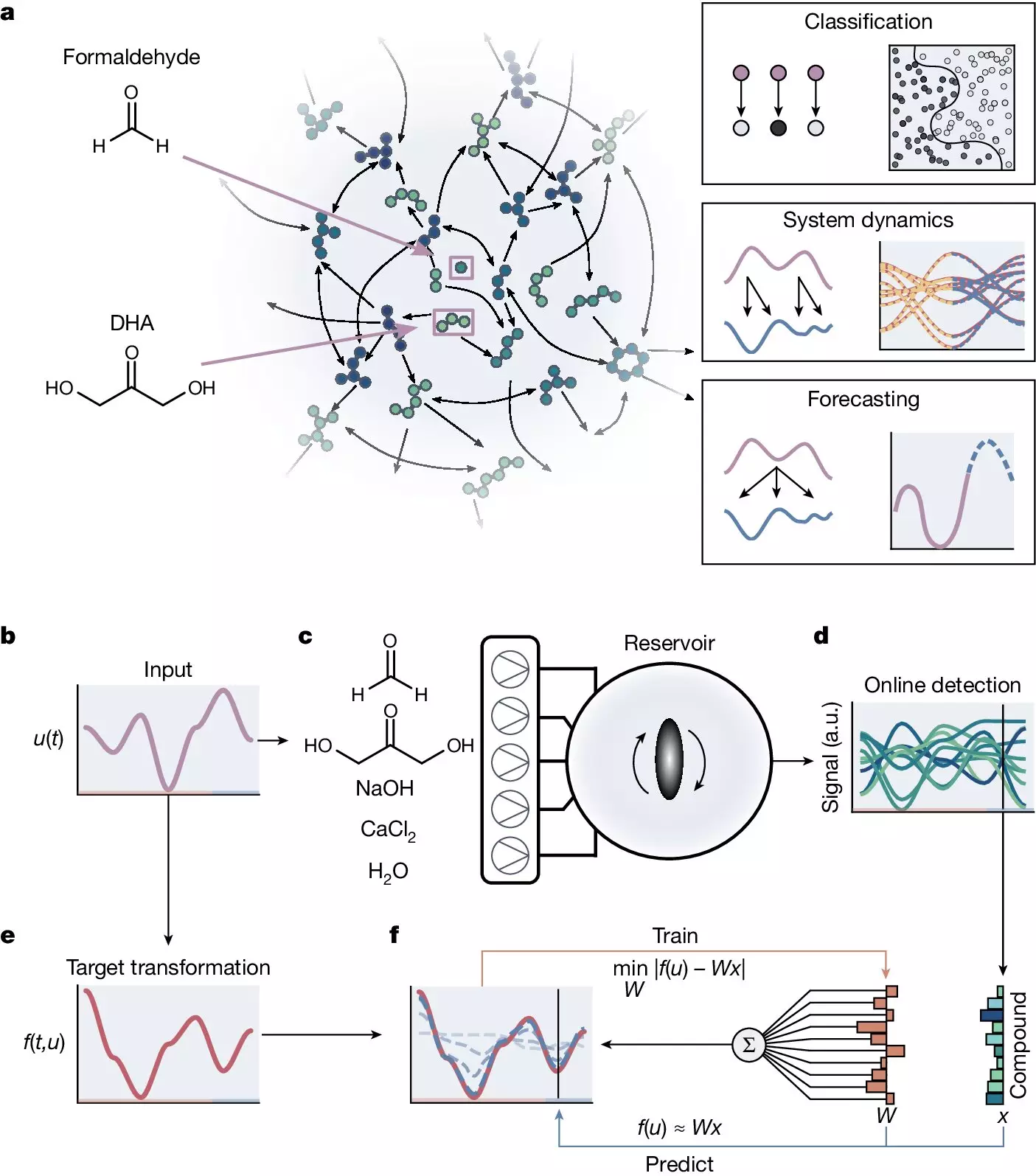
However, the road to implementing molecular computing is not without its obstacles. One of the primary challenges lies in the engineering and design aspects of these systems. Instead of attempting to engineer molecular systems to carry out specific computational tasks, Prof. Huck and his team are taking a different approach. They are exploring how naturally complex chemical systems can exhibit emergent computational properties. By studying the formose reaction, a chemical process that synthesizes sugars from formaldehyde, they aim to uncover the inherent computational capabilities of these systems.
The formose reaction stands out due to its unique properties. Unlike many other chemical reactions that follow a linear sequence, the formose reaction is a self-organizing reaction network with a highly non-linear topology. This network contains numerous positive and negative feedback loops, resulting in dynamic reactions that produce multiple intermediate compounds. This non-linear nature, coupled with the presence of feedback loops, gives rise to a diverse set of chemical species. The network’s self-organizing nature allows it to evolve and react to chemical inputs without external intervention, thereby producing a wide range of outputs.
To explore the computational capabilities of the formose reaction, the researchers utilized a continuous stirred tank reactor (CSTR). By controlling the input concentrations of four reactants – formaldehyde, dihydroxyacetone, sodium hydroxide, and calcium chloride – they were able to modulate the behavior of the reaction network. The output molecule was identified using a mass spectrometer, enabling the tracking of up to 10^6 molecules. This setup allowed the researchers to perform calculations, with the reactant concentrations serving as input values for the desired computations.
Before the system could effectively compute various tasks, it had to undergo a training phase. This involved finding a set of weights that would convert the mass spectrometer traces into the correct value of the computation. Once this linear regression problem was solved, the reservoir computer could compute the outcome for any new input based on the learned weights. This training step was crucial, as it enabled the reservoir to learn and predict how changes in input would affect the output.
Through their research, Prof. Huck and his team explored several computational tasks using the reservoir computer. They successfully executed nonlinear classification tasks, emulating Boolean logic gates and more complex classifications like XOR, checkers, circles, and sine functions. Furthermore, they were able to predict the behavior of a complex metabolic network model of E. coli and forecast future states of chaotic systems, such as the Lorenz attractor. The system exhibited short-term memory capabilities, retaining information about past inputs, and demonstrated the possibility of a fully chemical readout using colorimetric reactions.
The research on molecular computing holds promising implications for bridging the gap between artificial systems and the information processing capabilities of living cells. This new approach to molecular computing offers a scalable and flexible way to create autonomous chemical systems that can process information and respond to their environment without external electronic control. Looking ahead, Prof. Huck envisions embedding reservoir computing into chemical systems that can sense their surroundings, process information, and take appropriate action, potentially leading to exciting developments in neuromorphic computing and shedding light on the origins of life.

Leave a Reply