Proteins are fundamental components of cells, responsible for a wide range of crucial functions. Understanding their three-dimensional structures is essential for unraveling their roles in biological processes. In a groundbreaking development, the HUN-REN-ELTE Protein Modeling Research Group has introduced a novel mathematical method, known as LoCoHD (Local Composition Hellinger Distance), for comparing protein structures with remarkable precision. Unlike existing techniques that focus solely on atomic positions, LoCoHD incorporates crucial chemical information about the atoms, providing a comprehensive analysis of protein configurations.
The Complexity of Protein Structures
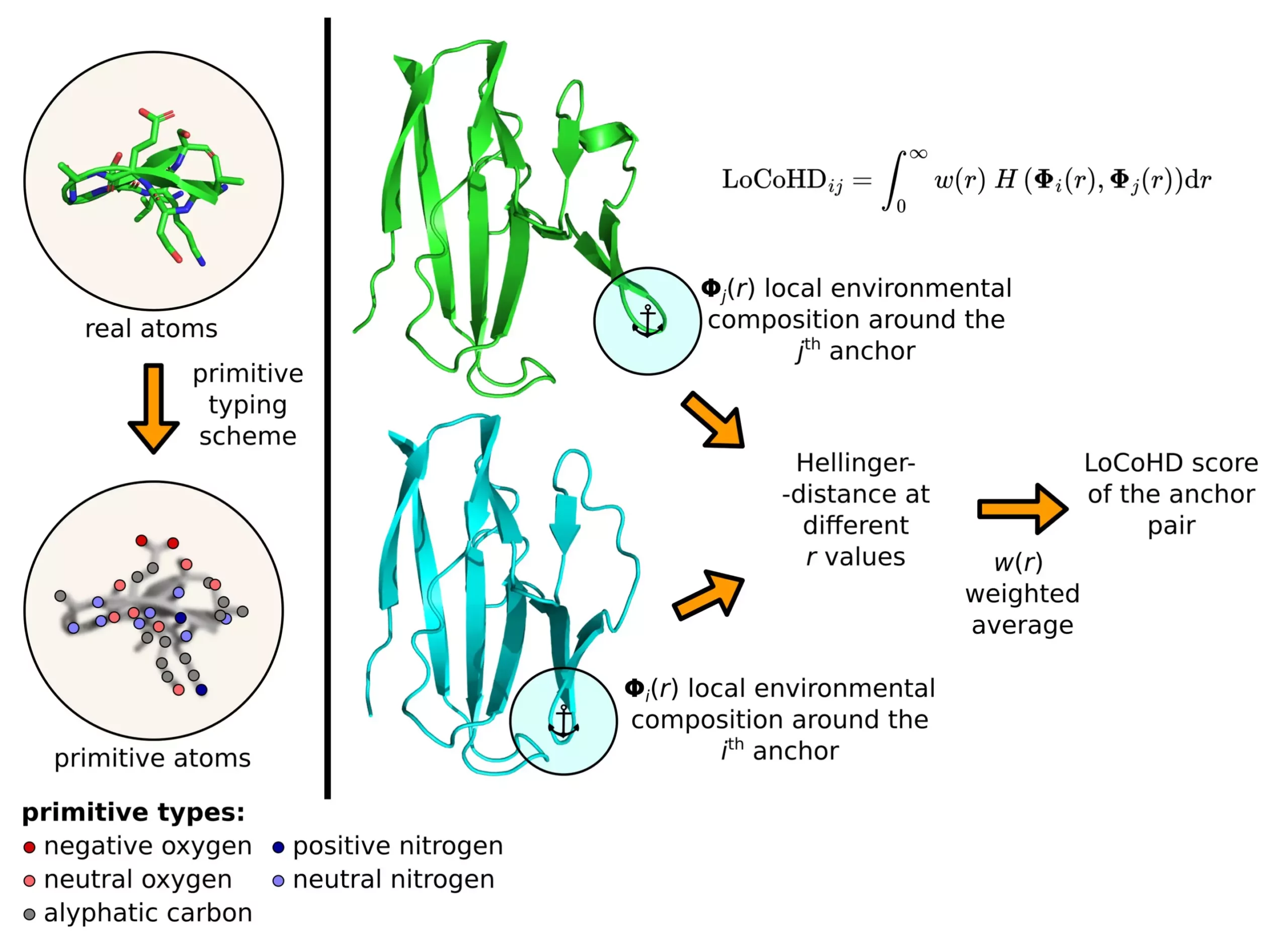
Proteins exhibit intricate three-dimensional arrangements that dictate their functionality within cells. Experimental methods such as X-ray crystallography, nuclear magnetic resonance spectroscopy, and cryo-electron microscopy have enabled scientists to determine the structures of numerous proteins. As the volume of available protein structure data continues to grow, the demand for advanced computational tools to analyze these intricate arrangements has escalated. LoCoHD, developed by Zsolt Fazekas and Dr. András Perczel’s research group, offers a sophisticated algorithmic approach to comparing local amino acid environments in proteins based on their chemical properties.
LoCoHD utilizes a multi-step protocol to generate a numerical metric representing the dissimilarity between protein structures. In the initial step, real atoms in the protein are converted into primitive atoms, each labeled with specific chemical characteristics. These primitive atoms serve as the basis for comparison, allowing the algorithm to evaluate structural differences effectively. By selecting anchor atoms as reference points for comparison, the method quantifies the dissimilarity between protein structures with unparalleled accuracy. The resulting metric can provide valuable insights into the structural nuances of proteins, shedding light on their biological functions.
Applicability in Protein Structure Prediction
The efficacy of LoCoHD extends beyond structural analysis, making it a valuable tool for protein structure prediction challenges such as CASP competitions. By incorporating the chemical composition of amino acid environments, the method offers a unique advantage in assessing modeled protein structures accurately. In the recent CASP14 competition, researchers utilized LoCoHD to compare various predicted protein structures, including those generated by cutting-edge techniques like AlphaFold2. Notably, the analysis of the SARS-CoV-2 virus protein ORF8 revealed substantial differences in amino acid interactions between experimental and predicted structures, highlighting the method’s precision in detecting structural variations.
In addition to static structure analysis, the researchers evaluated the applicability of LoCoHD in studying the dynamic behaviors of proteins. Through molecular simulations and structural ensemble data, the method successfully identified amino acids undergoing significant chemical environmental changes during protein motion. For instance, in the podocin protein critical for kidney function, LoCoHD pinpointed key amino acids influencing both structure and function. Similarly, the study of the HIV-1 capsid protein yielded insights into amino acid alterations crucial for viral envelope formation, underscoring the method’s versatility in investigating protein dynamics.
Implications for Disease Research and Drug Development
By enabling a deeper understanding of protein structures and dynamics, LoCoHD has the potential to advance research on disease-causing pathogens and facilitate drug development. Through precise analysis of amino acid environments and structural variations, scientists can gain novel insights into the mechanisms underlying severe diseases. This knowledge paves the way for the development of targeted therapeutics and interventions designed to combat pathogenic proteins effectively. With the continued refinement and application of advanced computational methods like LoCoHD, researchers are poised to unlock new frontiers in protein structure analysis and biomedical research.

Leave a Reply