In the complex world of cell biology, ribosomes serve as essential machines responsible for synthesizing proteins, the building blocks of life. Recent advancements in computational biology are shedding light on the intricate processes occurring within these cellular powerhouses. Researchers at the University of Tsukuba have made significant strides by developing an innovative model that simulates the ribosome’s internal environment. This endeavor aims to deepen our understanding of protein synthesis, particularly focusing on how proteins begin to fold into functional forms while still inside the ribosomal tunnel.
Understanding the Ribosome Tunnel
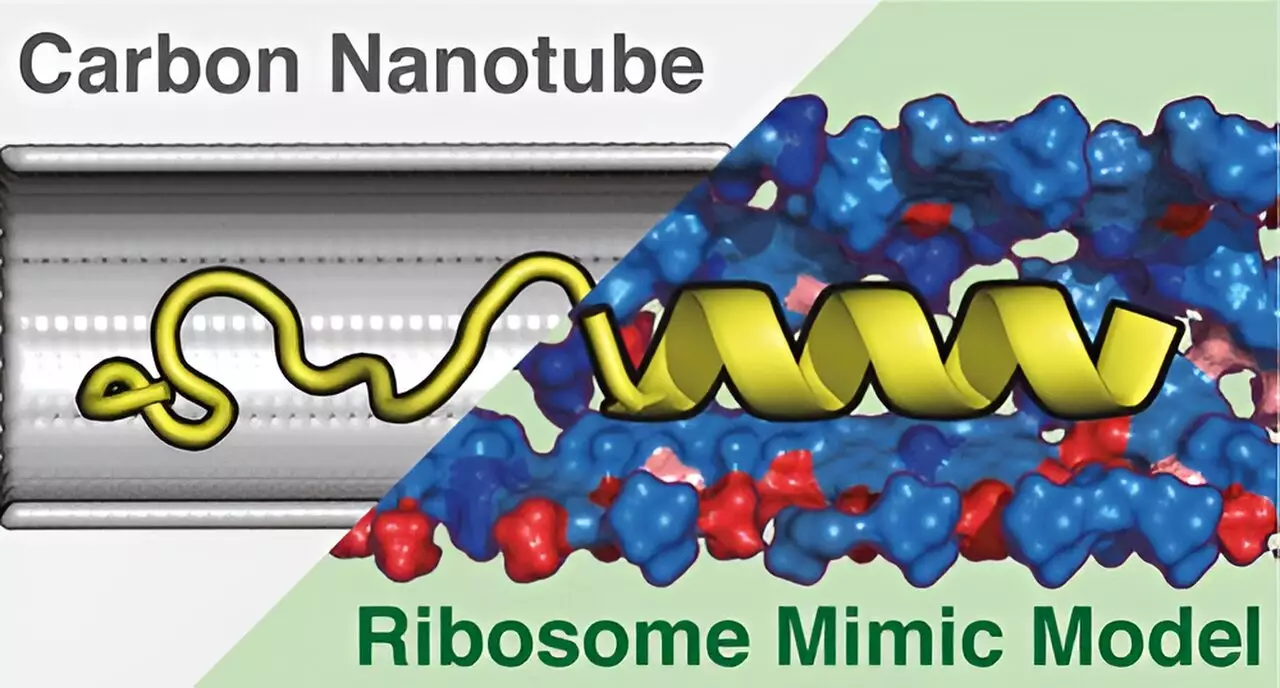
The ribosome tunnel is a specialized tubular channel that facilitates the exit of nascent proteins. Historically, it was believed that proteins emerge fully formed; however, the latest studies suggest a fascinating possibility: proteins may start folding into their three-dimensional structures while they navigate through this tunnel. This unfolding mystery raised a crucial question: how do the chemical properties and structural characteristics of the ribosome tunnel influence protein folding? Addressing this question required a more refined model to study these dynamics.
To tackle these complexities, the research team created the Ribosome Environment Mimicking Model (REMM). This cylindrical construct not only replicates the inner diameter of ribosomal tunnels but also incorporates key chemical properties critical for protein interaction and folding. Unlike conventional models, such as carbon nanotubes (CNT), which only account for the diameter without the necessary chemical diversity, REMM stands out for its comprehensive approach. The decision to integrate both structural and chemical parameters makes this model a significant advancement in the field.
Employing molecular dynamics simulations, the researchers analyzed how proteins interacted within both the REMM and CNT models. The results were striking. The REMM showed a remarkable ability to replicate experimentally observed protein structures more effectively than the CNT model. This enhancement is attributed to the chemical diversity provided by the REMM, which more closely mirrors the biological environment of the ribosomal tunnel. This finding underscores the importance of chemical interactions in the folding process of proteins and highlights the limitations of previous models that failed to incorporate these factors.
Implications for Future Research
The implications of this research are profound. With further refinement, the REMM has the potential to revolutionize our understanding of protein conformations in live cells. By offering insights into how proteins are synthesized and folded in real-time, this model could pave the way for significant advancements in drug design and protein engineering. As scientists strive to unlock the enigma of protein behavior at the molecular level, tools like REMM will be crucial in bridging the gap between theoretical knowledge and empirical observations.
The study conducted by the University of Tsukuba illustrates a significant leap forward in the comprehension of protein synthesis and folding mechanisms. As we continue to explore the complexities of ribosomes and their utility in understanding life’s processes, models like REMM will undoubtedly play a pivotal role in shaping future discoveries. The journey toward fully deciphering the intricacies of protein synthesis promises to enrich our biological insights and encourage groundbreaking innovations in various scientific fields.

Leave a Reply