The understanding of histones has traditionally revolved around multi-cellular organisms, where these proteins play a crucial role in the organization and compaction of DNA. Recent breakthroughs, however, underscore that even single-celled life forms such as bacteria and archaea host a rich diversity of histone proteins. A groundbreaking study conducted by Samuel Schwab and his team at the Leiden Institute of Chemistry has enhanced our comprehension of these proteins, revealing an astonishing variety of structural forms and potential functions that challenge previous assumptions about cell biology.
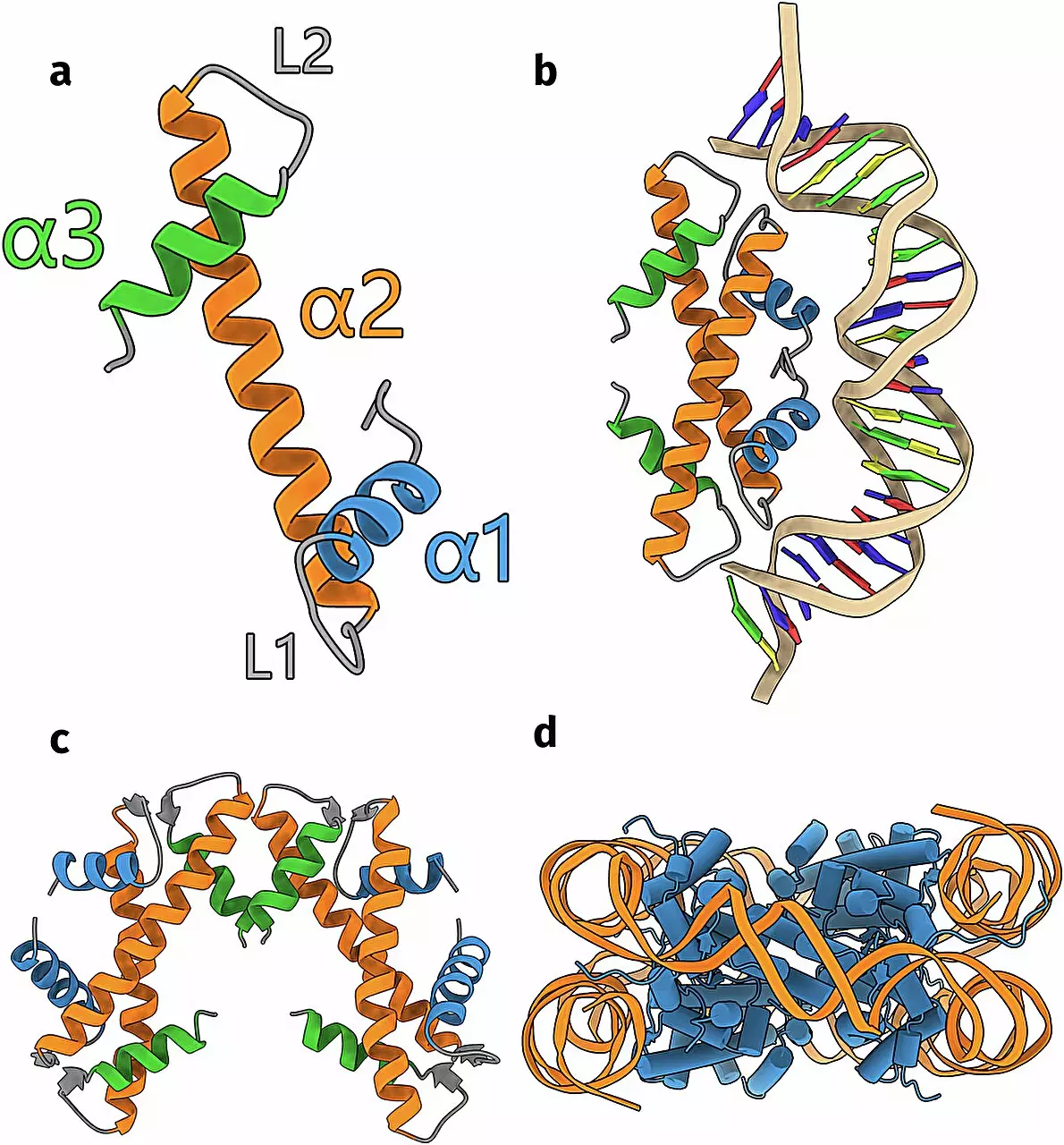
For many years, researchers were under the impression that histones were exclusive to higher-order life forms—essentially, complex organisms. It was only after dedicated investigations into archaea and bacteria that scientists came to recognize these simple, single-celled entities also utilize histones to manage their genetic material. Schwab’s research expands the notion that the diversity of histones is not only present but remarkably extensive, encompassing at least seventeen distinct groups, each exhibiting unique characteristics. This finding challenges conventional wisdom by elucidating the mechanisms through which even the simplest organisms manage DNA within their cells.
At the center of this research is the utilization of artificial intelligence, particularly the innovative AlphaFold algorithm. Schwab’s team leveraged this cutting-edge technology to predict the three-dimensional structures of histones based on DNA sequences derived from a vast protein database. The ability of AlphaFold to accurately model protein structures has provided researchers with a powerful tool, enabling them to explore regions of biology that were previously difficult to investigate due to technological limitations. Schwab’s research, informed by this AI approach, carried an expansive breadth, analyzing over 6,000 distinct DNA sequences potentially related to uncharacterized histones.
The integration of computational predictions with laboratory experiments is a hallmark of modern scientific inquiry, and Schwab’s study is no exception. After the team employed AlphaFold to predict histone structures, they endeavored to validate these models through experimental techniques. Their efforts were rewarded when they successfully elucidated the structure of a new group of histones, finding the results strikingly aligned with the computational predictions. This synchronicity between computational biology and empirical research underscores the reliability of AI-driven predictions, reinforcing the burgeoning relationship between technology and biological science.
Delving beyond mere structure, this research has opened new avenues for understanding the functional roles of histones within single-celled organisms. Traditional models primarily considered histones as components that wrap DNA into nucleosomes. Both Schwab and his supervisor, Remus Dame, contend that this view is overly simplistic. Their findings suggest that some histones exhibit an entirely different dynamic—potentially binding to cellular membranes instead of DNA—hinting at alternative roles in regulating cellular processes. This revelation prompts a reevaluation of how we interpret the organization of genetic material and the multifaceted interactions between proteins, DNA, and other cellular constituents.
As Schwab articulates, this investigation contributes not only to a more nuanced appreciation for the diversity of histones but also to the evolutionary implications surrounding DNA organization. Understanding these variations in protein structures helps researchers infer evolutionary relationships and adaptations across different life forms. Moreover, these insights furnish a more profound comprehension of intracellular dynamics, laying groundwork for future studies that may elucidate biochemical pathways previously obscured by a lack of knowledge regarding histone functionality.
The journey to fully unlock the mysteries associated with histones in archaea and bacteria is only beginning. Schwab emphasizes that substantial gaps in knowledge remain, particularly regarding the mechanistic roles these proteins undertake. As future research endeavors focus on these lesser-understood elements of cellular biology, the potential applications of this knowledge could span multiple fields from genetics to biotechnology, fostering advancements that hinge on a deeper understanding of life’s most fundamental units. Schwab’s study elegantly illustrates the sophistication of cellular functions in single-celled organisms and opens the door for further exploration into the intricacies of histone biology.

Leave a Reply