The study conducted by a research team has introduced a groundbreaking technique that could potentially revolutionize the way scientists identify novel targets for cancer therapies. In the world of biology, precision is key, particularly when it comes to the editing of RNA messages within living cells. This editing process plays a crucial role in maintaining overall cell health and preventing diseases such as cancer. However, until now, observing this intricate editing process in detail has been challenging. This recent development by a team of WashU chemists, spearheaded by graduate student Alex Quillin, has opened up new possibilities for unraveling the mysteries of RNA editing.
RNA editing is a fundamental process that allows cells to function properly, yet it often goes awry in cases of disease. Despite its importance, there is still much to learn about the timing, location, and regulation of RNA editing within cells. The most common type of RNA edit involves converting adenosine molecules into inosine using a specialized enzyme. This conversion is essential for distinguishing between normal RNA and foreign RNA, enabling cells to detect and respond to potential threats. Without these edits, cells may fail to trigger the immune system when necessary. Previous methods of tracking RNA edits lacked the precision needed to pinpoint their exact locations within cells, hindering researchers’ ability to fully understand the process.
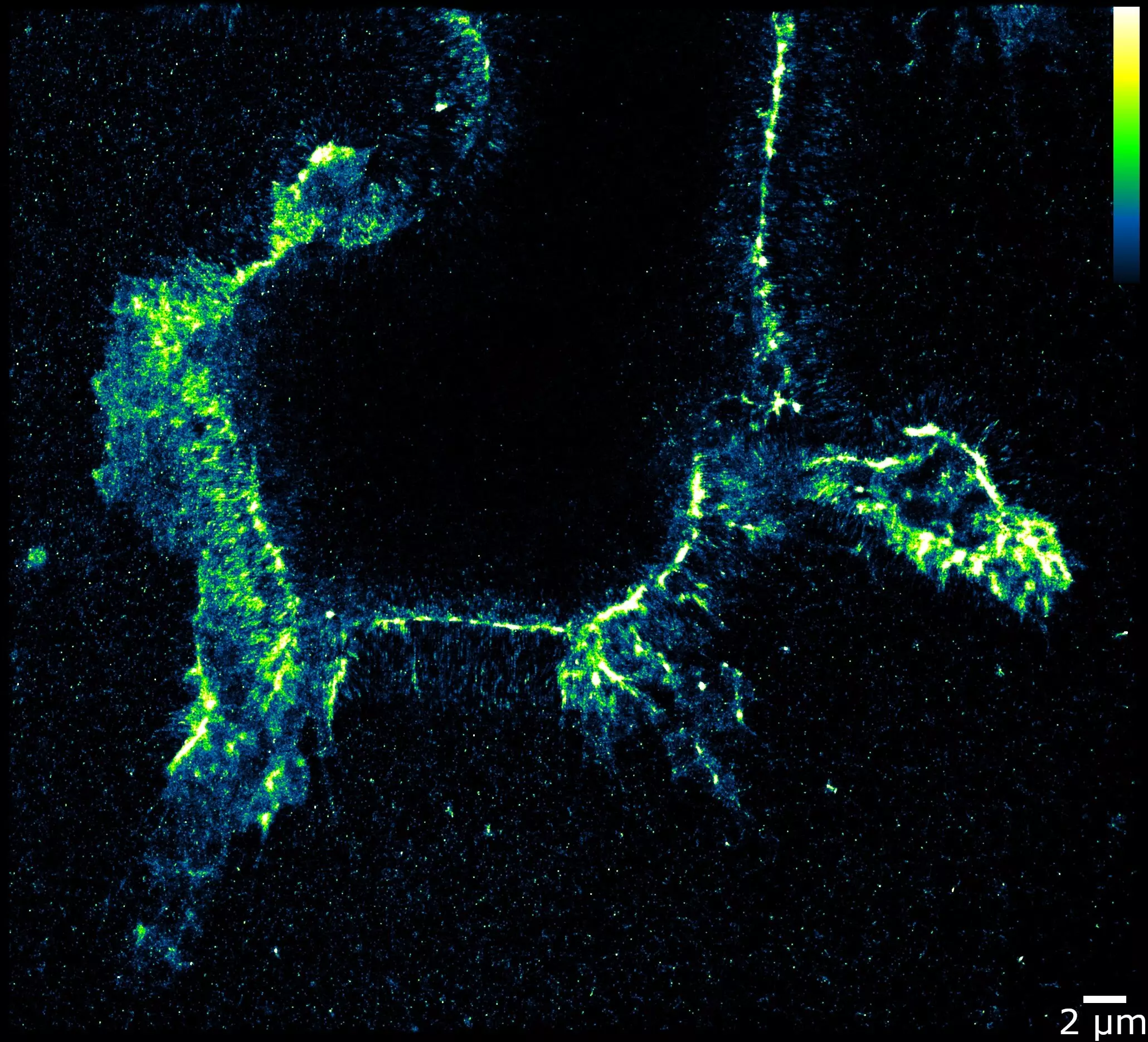
The EndoVIA test developed by the WashU team represents a significant advancement in RNA editing research. By repurposing an enzyme that targets edited RNAs and modifying the conditions under which it functions, researchers are now able to visualize individual RNA edits within cells. This innovative approach allows for the precise tracking of editing patterns, shedding light on the origins of various diseases. The use of fluorescent antibodies to tag the enzyme has enabled researchers to observe the editing process with unprecedented clarity, offering valuable insights into the intricacies of RNA editing.
The implications of the EndoVIA technique go beyond basic research, holding the potential to impact cancer therapy worldwide. Some cancers have been linked to abnormalities in RNA editing, making it crucial to identify and understand these patterns. For example, breast cancer cells have been found to exhibit excessive editing, while kidney cancer cells tend to have insufficient edits. Through their study of healthy and cancerous kidney cells, Quillin and colleagues were able to identify distinct editing patterns near cell membranes that were missing in cancerous cells. Although the significance of these differences remains unclear, this discovery paves the way for future investigations into their biological implications.
The development of the EndoVIA technique marks a significant milestone in RNA editing research, offering researchers a powerful tool for studying the intricacies of cellular processes. The ability to visualize and track individual RNA edits within cells provides valuable insights into the mechanisms underlying diseases such as cancer. By pinpointing specific editing patterns associated with different types of cancer, scientists can potentially identify novel targets for therapeutic interventions. The groundbreaking work carried out by the WashU team has the potential to shape the future of cancer therapy and advance our understanding of RNA editing in cellular health.

Leave a Reply